Research and citations
Since a few months now, the Aalto University CRIS back-end has served the research active personnel and other inclined staff. Since last week, the front-end site was opened to the public. One of the most interesting aspects of the CRIS is that it offers, in one place, a digital dossier of one’s publications, together with their metrics.
The de facto metrics in science is the number of citations, made available by the two leading commercial indexing services, Web of Science and Scopus. The Pure CRIS system can talk with both via an API, and return the citation count. For the discussion though, Pure needs to know the exact topic of the talk, the unique identification number that the publication has got in these respective indexing services. In Web of Science, the ID is known as UT or WOS. In Scopus, just ID.
In pre-CRIS times, publication metadata was stored in legacy registers. In these, external IDs were an unknown entity. When old metadata was transformed to the Pure data model and imported, these new records were all missing both UT and Scopus ID. Manually copy-pasting them all would be a daunting task, bordering impossible. A programmatic solution offers some remedy here: IDs can be imported to Pure with a ready-made job. For that, you also need the internal record ID of Pure. The input file of the job is a simple CSV with a pair of Pure ID and external ID per row. The challenge is, how to find the matching pairs.
Getting data
Within the Web of Science GUI, you can make an advanced Boolean affiliation search. What this means is that you need to be familiar with all historic name variants of your organization, added with known false positives that you better exclude. I’m not copying our search here, but I can assure you: it is a long and winding string. The result set, filtered by publication years 2010-2016 (the Aalto University period so far) is over 20.000 items. Aalto University outputs roughly 4000 publications yearly.
In what format to download the WoS result data set? There are several possibilities. The most used one I assume, is tabular-limited Windows aka Excel. There is a caveat though. If the character length of some WoS field value exceeds the Excel cell limit, the tabular data is garbled from that point onwards. This happens quite often with the Author field where the names of all contributing authors are concatenated. However, the WoS plain text format doesn’t have this problem, but the output needs parsing before it’s usable. In the exported file, fields are denoted with a two-character label or tag. Fields can span over multiple lines. For this exercise, I’m mostly interested in title (TI), name (SO), ISSN (SN, EI), and article identifier (UT).
As a CRIS (Pure) back-end user, to fetch tabular data out of the system, you have a number of options at the moment:
- Filtering from the GUI. Quick and easy but doesn’t provide all fields, e.g. record ID
- Reporting module. Flexible but asks for a few rounds of trial & error, plus the Administrator role if fields you need are not available by default in which case you need to tweak the report configurations. Minimum requirement is the Reporter role. However, you can save your report specifications, and even import/export them in XML, which is nice
- REST API. For my simple reporting needs, in its present sate, the API would require too much effort in constructing the right query, and parsing the result
Of course there is also the SQL query option, but for that, you need to be familiar with the database schema, and be granted all necessary access rights.
With the Pure Reporting module, filtering by year, and finding publication title, subtitle, ISSN and ID is relatively easy. From the output formats, I choose Excel. This and the parsed WoS data I will then read into R, and start matching.
To parse the WoS data, I chose the Unix tool awk.
But before parsing, data download. For this, you need paper and pen.
There is an export limit of 500 records at WoS, so you have to select, click and save 25 times to get all the 20K+ result set. To keep a manual record of the operation is a must, otherwise you’ll end up exporting some items several times, or skipping others.
Parsing
When all WoS data is with you, the Bash shell script below does the work. Note that I’ve added newlines in longer commands for clarity. You may also notice, that I had some troubles in defining appropiate if clauses in awk, to handle ISSNs. By brute force parsing, I circumvented the obstacle, and made housekeeping afterwards.
#!/bin/bash
# Delete output file from previous run
rm parsed.txt
# Rename the first WoS output file
mv savedrecs.txt savedrecs\(0\).txt
# Loop over all exported files and filter relevant fields
for i in {0..25}
do
awk 'BEGIN {ORS="|"}; # ORS = output field separator
/^TI/,/^LA/ {print $0}; # match boundaries, to get the SO field that's in between there
/^DT/ {print $0};
/^SN/ {print $0};
/^EI/ {print $0};
/^UT/ {printf "%s\n",$0}' savedrecs\($i\).txt >> parsed.txt
done
# Delete labels, LA field, occasional SE field, and extra white space resulting
# from fields with multiple lines
sed -i 's/^TI //g;
s/| / /g;
s/|SO /|/g;
s/|LA [^|]*//g;
s/|DT /|/g;
s/|SN /|/g;
s/|UT /|/g;
s/|SE [^|]*//g' parsed.txt
# Filter out rows without a second ISSN
grep -v '|EI ' parsed.txt > noEI.txt
# Filter out rows with a second ISSN
grep '|EI ' parsed.txt > yesEI.txt
# Add and empty field to those without, just before the last field
sed -i 's/|WOS/||WOS/g' noEI.txt
# Concat these two files
cat noEI.txt yesEI.txt > parsed_all.csv
# Delete the label of the second ISSN
sed -i 's/|EI /|/g' parsed_all.csv
# If, before '||', which denotes a non-existing 2nd ISSN, there is no match for an ISSN
# (so there wasn't any in data, meaning the publication is not an article),
# add an empty field
sed -i '/|[0-9][0-9][0-9][0-9]\-[0-9][0-9][0-9][0-9X]||/! s/||/|||/g' parsed_all.csv
After a few seconds, the parsed_all.csv includes rows like the following three ones:
Modulation Instability and Phase-Shifted Fermi-Pasta-Ulam Recurrence|SCIENTIFIC REPORTS|Article|2045-2322||WOS:000379981100001 Real World Optimal UPFC Placement and its Impact on Reliability|RECENT ADVANCES IN ENERGY AND ENVIRONMENT SE Energy and Environmental Engineering Series|Proceedings Paper|||WOS:000276787500010 Deposition Order Controls the First Stages of a Metal-Organic Coordination Network on an Insulator Surface|JOURNAL OF PHYSICAL CHEMISTRY C|Article|1932-7447||WOS:000379990400030
Processing in R
There are several R packages for reading and writing Excel. XLConnect has served me well on Ubuntu Linux. However, on my Windows laptop at home, its Java dependencies have been a minor headache lately.
library(XLConnect)
wb <- loadWorkbook("puredata20160909.xls")
puredata <- readWorksheet(wb, sheet = "Research Output")
puredata <- puredata[, c("Title.of.the.contribution.in.original.language.1", "Subtitle.of.the.contribution.in.original.language.2",
"Journal...Journal.3", "Id.4", "Journal...ISSN...ISSN.5")]
names(puredata) <- c("title", "subtitle", "journal", "id", "issn")
# Paste title and subtitle, if there
puredata$title <- ifelse(!is.na(puredata$subtitle), paste(puredata$title, puredata$subtitle), puredata$title)
puredata <- puredata[, c("title", "journal", "id", "issn")]
WoS data file import:
wosdata <- read.csv("parsed_all.txt", stringsAsFactors = F, header = F, sep = "|", quote = "", row.names = NULL)
names(wosdata) <- c("title", "journal", "type", "issn", "issn2", "ut")
Before attempting to do any string matching though, some harmonization is necessary: all chars to lowercase, remove punctuation and articles, etc. Without going into details, here is my clean function. The gsub clauses are intentionally separated one to each for easier reading. Also, I haven’t used any character class but defined characters one by one.
clean <- function(dataset) {
# Journal
dataset$journal <- ifelse(!is.na(dataset$journal), tolower(dataset$journal), dataset$journal)
# WOS
if ( "ut" %in% names(dataset) ) {
dataset$ut <- gsub("WOS:", "", dataset$ut)
}
# Title
dataset$title <- tolower(dataset$title)
dataset$title <- gsub(":", " ", dataset$title)
dataset$title <- gsub(",", " ", dataset$title)
dataset$title <- gsub("-", " ", dataset$title)
dataset$title <- gsub("%", " ", dataset$title)
dataset$title <- gsub('\\"', ' ', dataset$title)
dataset$title <- gsub('\\?', ' ', dataset$title)
dataset$title <- gsub("\\([^)]+\\)", " ", dataset$title)
dataset$title <- gsub(" the ", " ", dataset$title)
dataset$title <- gsub("^[Tt]he ", "", dataset$title)
dataset$title <- gsub(" an ", " ", dataset$title)
dataset$title <- gsub(" a ", " ", dataset$title)
dataset$title <- gsub("^a ", "", dataset$title)
dataset$title <- gsub(" on ", " ", dataset$title)
dataset$title <- gsub(" of ", " ", dataset$title)
dataset$title <- gsub(" for ", " ", dataset$title)
dataset$title <- gsub(" in ", " ", dataset$title)
dataset$title <- gsub(" by ", " ", dataset$title)
dataset$title <- gsub(" non([^\\s])", " non \\1", dataset$title)
dataset$title <- gsub("^[[:space:]]*", "" , dataset$title)
dataset$title <- gsub("[[:space:]]*$", "" , dataset$title)
# ISSN
dataset$issn <- gsub("-", "" , dataset$issn)
dataset$issn <- gsub("[[:space:]]*$", "" , dataset$issn)
dataset$issn <- gsub("^[[:space:]]*", "" , dataset$issn)
# Second ISSN
if ( "issn2" %in% names(dataset) ) {
dataset$issn2 <- gsub("-", "" , dataset$issn2)
dataset$issn2 <- gsub("[[:space:]]*$", "" , dataset$issn2)
dataset$issn2 <- gsub("^[[:space:]]*", "" , dataset$issn2)
}
return(dataset)
}
Match by joining
The R dplyr package is indispensable for all kind of data manipulation. One of the join types it supports (familiar from SQL), is left_join, which
returns all rows from x, and all columns from x and y. Rows in x with no match in y will have NA values in the new columns. If there are multiple matches between x and y, all combinations of the matches are returned.
So here I am saying to R:Compare my two data frames by title and ISSN, as they are given. Please.
library(dplyr)
# Join with the first ISSN
joined <- left_join(puredata, wosdata, by=c("title"="title", "issn"="issn"))
# and then with the second ISSN
joined2 <- left_join(puredata, wosdata, by=c("title"="title", "issn"="issn2"))
found <- joined[!is.na(joined$ut),]
found2 <- joined2[!is.na(joined2$ut),]
names(found) <- c("title", "journal", "id", "issn", "journal2", "type", "issn2", "ut")
names(found2) <- c("title", "journal", "id", "issn", "journal2", "type", "issn2", "ut")
allfound <- rbind(found, found2)
This way, roughly 55% of CRIS articles, published between 2010 and 2016, found a match – and the needed WOS/UT. Then, similarly, I matched conference publications with title, i.e. items without a corresponding ISSN. Here, the match rate was even better, 73%, although the number of publications was not that big.
However, only one typo in title or ISSN – and no match could happen. Thanks to my smart coworkers, I was made aware of the Levenshtein distance. In R, this algorithm is implemented in few packages.
Fuzzier match
I started with the adist function from the utils package that is normally loaded automatically when you start R. The code examples from Fuzzy String Matching – a survival skill to tackle unstructured information were of great help. The initial tests with a smallish sample looked promising.
Pretty soon it became obvious though, that it would take too much time to process my whole data in this manner. If 1000 x 1000 rows took 15 minutes, 6000 x 12000 rows would take probably ten times that, or maybe even more. What I needed was to run the R code in parallel.
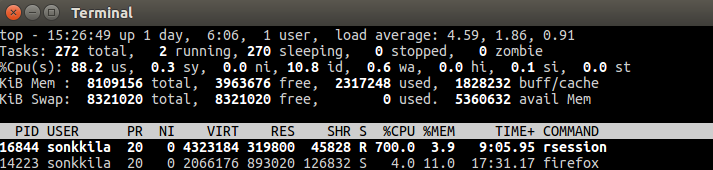
My Linux machine at work runs a 64-bit Ubuntu 16.04, with 7.7 GB RAM and 2.67 GHz Intel Xeon processors, ” x 8″, as the System settings concludes. I’m not sure if 8 here refers to the number of cores or threads. Anyway, if only my code could be parallelized, I could let the computer or other higher mind to decide how to core-source the job in the most efficient way.
Luckily, there is the stringdist package by Mark van der Loo that does just that. By default, it parallelizes your work between all cores you’ve got, minus one. This is common sense, because while your code is running, you may want to do something else with you machine, like watch the output of the top command.
Here I am running the amatch function from the package, trying to match those article titles that were not matched in the join. The method argument value lv stands for Levenshtein distance, as in R’s native adist. maxDist controls how many edits from the first string to the second is allowed for a match. The last argument defines the output in nomatch cases.
system.time(matched <- amatch(notfound_articles$title, wosdata_articles$title, method="lv", maxDist = 5, nomatch = 0))
Look: %CPU 700!
I haven’t tried to change the default nthread argument value, so I don’t know if my run time of roughly 6 minutes could be beaten. Nevertheless, six minutes (363.748 milliseconds) is just fantastic.
user system elapsed
2275.980 0.648 363.748
In the matched integer vector, I had now, for every row from notfound_articles data frame, either a 0 or a row number from wosdata_articles. Among the first twenty one rows, there are already two matches.
>matched[1:20] [1] 0 0 0 0 0 55 0 0 0 0 0 0 0 0 0 0 1 0 0 0
All I needed to do, was to gather the IDs of the matching strings, and output. For the sake of quality control, I’m picking up also the strings.
N <- length(matched)
pure <- character(N)
pureid <- character(N)
wos <- character(N)
ut <- character(N)
for (x in 1:N) {
pure[x] <- notfound_articles$title[x]
pureid[x] <- notfound_articles$id[x]
wos[x] <- ifelse(matched[x] != 0, wosdata_articles[matched[x], "title"], "NA")
ut[x] <- ifelse(matched[x] != 0, wosdata_articles[matched[x], "ut"], "NA")
}
df <- data.frame(pure, pureid, wos, ut)
df <- df[df$wos!='NA',c("pureid", "ut")]
names(df) <- c("PureID", "ExternalID")
write.csv(df, paste0("pure_wos_fuzzy_articles", Sys.Date(), ".csv"), row.names = F)
Then, the same procedure for the conference output, and I was done.
All in all, thanks to stringdist and other brilliant stuff by the R community, I could find a matching WoS ID to 67.7% of our CRIS publications from the Aalto era.
To repeat the same process for Scopus means to build another parser, because the export format is different. The rest should follow similar lines.
EDIT 15.9.2016: Made corrections to the shell script.